The challenge from the KDD Cup this year was to use their data relating to student enrollment in online MOOCs to predict who would drop out vs who would stay.
The short story is that using H2O and a lot of my free time, I trained several hundred GBM models looking for the final one which eventually got me an AUC score of 0.88127 on the KDD Cup leaderboard and at the time of this writing landed me in 120th place. My score is 2.6% away from 1st place, but there are 119 people above me!
Here are the main characters of this story:
It started with my obsessive drive to find an analytics project to work on. I happened upon the KDD Cup 2015 competition and decided to give it a go. It had the characteristics of a project that I wanted to get into:
1) I could use it to practice my SQL skills
2) The data set was of a moderate size (training table was 120,542 records, log info table was 8,151,053 records!)
3) It looked like it would require some feature engineering
4) I like predictive modeling competitions 
Once I had loaded up the data into a mariadb database, I had to come to decisions about how I would use the info in each table. Following were my thought processes for each table:
enrollment_train / enrollment_test
Columns: enrollment_id, username, course_id
Simply put, from this table I extracted the number of courses each student (username) was enrolled in, and also the number of students enrolled in each course (course_id).
log_train / log_test
Columns: enrollment_id, tstamp, source, logged_event, object
There were a few items of information that I decided to extract from this table:
1) Number of times each particular event was logged for every enrollment_id
2) Average timestamp for each event for each enrollment_id
3) Min and Max timestamp for each event for each enrollment_id
4) Total time elapsed from the first to the last instance of each event for each enrollment_id
5) Overall average timestamp for each enrollment_id
Contrary to what you might think, the object field does not seem to link up with the object table.
object
Columns: course_id, module_id, category, children, tstart
From this table I extracted a count of course components by course_id and also the number of ‘children’ per course_id. I assume these are relational references but am not sure what in the data set these child IDs refer to.
truth_train
Columns: enrollment_id, dropped_out
I didn’t extract anything special out of this table, but used it as the table to which all other SQL views that I had created were linked.
If you’d like to see the SQL code I used to prepare the tables, views, and the final output table I used to train the model, see my github repo for this project.
Import into R and Feature Engineering
Once I imported the data into R through RODBC, you’ll see in the code that my feature engineering was essentially a desperate fishing expedition where I tried a whole lot of stuff. I didn’t even end up using everything that I had engineered through my R code, but as my final model included 35 variables, I wasn’t suffering any severe lack! If you download the KDD Cup 2015 data and are having a look around, feel free to let me know if I’ve missed any important variables!
H2O, Model Tuning, and Training of The Final Model
This is the part where I managed to train hundreds of models! I don’t think this would have been feasible just using plain R on my computer alone (I have 8GB of RAM and an 8 core AMD processor). For these tasks I turned to H2O. For those who don’t know, H2O is a java based analytical interface for cloud computing that is frankly very easy and beneficial to set up when all you have at your disposal is one computer. I say beneficial for one reason: my computer chokes when trying to train ensemble models on even moderate sized data sets. Through H2O, I’m able to get it done without watching the RAM meter on my system monitor shoot all the way up to full capacity!! What you’ll notice in my R code is that R is able to interface with H2O in such a way that once I passed the dataframe with the training data to H2O, it was H2O that handled the modeling from there, and sends info back to R when available or requested (e.g. while you’re training a model, it gives you a cute text-based progress bar automatically!). More on this soon.
Before I show some results, I want to talk about my model tuning algorithm. Let’s look at the relevant code, then I’ll break it down verbally.
ntree = seq(100,500,100)
balance_class = c(TRUE,FALSE)
learn_rate = seq(.05,.4,.05)
parameters = list(ntree = c(), balance_class = c(), learn_rate = c(), r2 = c(), min.r2 = c(), max.r2 = c(), acc = c(), min.acc = c(), max.acc = c(), AUC = c(), min.AUC = c(), max.AUC = c())
n = 1
mooc.hex = as.h2o(localH2O, mooc[,c("enrollment_id","dropped_out_factor",x.names)])
for (trees in ntree) {
for (c in balance_class) {
for (rate in learn_rate) {
r2.temp = c(NA,NA,NA)
acc.temp = c(NA,NA,NA)
auc.temp = c(NA,NA,NA)
for (i in 1:3) {
mooc.hex.split = h2o.splitFrame(mooc.hex, ratios=.8)
train.gbm = h2o.gbm(x = x.names, y = "dropped_out_factor", training_frame = mooc.hex.split[[1]],
validation_frame = mooc.hex.split[[2]], ntrees = trees, balance_classes = c, learn_rate = rate)
r2.temp[i] = train.gbm@model$validation_metrics@metrics$r2
acc.temp[i] = train.gbm@model$validation_metrics@metrics$max_criteria_and_metric_scores[4,3]
auc.temp[i] = train.gbm@model$validation_metrics@metrics$AUC
}
parameters$ntree[n] = trees
parameters$balance_class[n] = c
parameters$learn_rate[n] = rate
parameters$r2[n] = mean(r2.temp)
parameters$min.r2[n] = min(r2.temp)
parameters$max.r2[n] = max(r2.temp)
parameters$acc[n] = mean(acc.temp)
parameters$min.acc[n] = min(acc.temp)
parameters$max.acc[n] = max(acc.temp)
parameters$AUC[n] = mean(auc.temp)
parameters$min.AUC[n] = min(auc.temp)
parameters$max.AUC[n] = max(auc.temp)
n = n+1
}
}
}
parameters.df = data.frame(parameters)
parameters.df[which.max(parameters.df$AUC),]
The model that I decided to use is my usual favourite, gradient boosting machines (h2o.gbm is the function you use to train a gbm model through H2O). As such, the 3 hyperparameters which I chose to vary and evaluate in the model tuning process were number of trees, whether or not to balance the outcome classes through over/undersampling, and the learning rate. As you can see above, I wanted to try out numerous values for each hyperparameter, making 5 values for number of trees, 2 values for balance classes, and 8 values for learning rate, totalling 80 possible combinations of all 3 hyperparameter values together. Furthermore, I wanted to try out each combination of hyperparemeter values on 3 random samples of the training data. So, 3 samples of each one of 80 combinations is equal to 240 models trained and validated with the aim of selecting the one with the best area under the curve (AUC). As you can see, each time I trained a model, I saved and summarised the validation stats in a growing list which I ultimately converted to a data.frame and called called parameters.df
The best hyperparameters, according to these validation stats which I collected, are:
– ntree = 500
– balance_class = FALSE
– learn_rate = .05
You can see a very nice summary of how validation set performance changed depending on the values of all of these parameters in the image below (the FALSE and TRUE over the two facets refer to the balance_class values.
Have a look at my validation data model summary output from the H2O package below:
H2OBinomialMetrics: gbm
** Reported on validation data. **
MSE: 0.06046745
R^2: 0.102748
LogLoss: 0.2263847
AUC: 0.7542866
Gini: 0.5085732
Confusion Matrix for F1-optimal threshold:
dropped out stayed Error Rate
dropped out 21051 1306 0.058416 =1306/22357
stayed 1176 576 0.671233 =1176/1752
Totals 22227 1882 0.102949 =2482/24109
Maximum Metrics:
metric threshold value idx
1 max f1 0.170555 0.317006 198.000000
2 max f2 0.079938 0.399238 282.000000
3 max f0point5 0.302693 0.343008 134.000000
4 max accuracy 0.612984 0.929321 48.000000
5 max precision 0.982246 1.000000 0.000000
6 max absolute_MCC 0.170555 0.261609 198.000000
7 max min_per_class_accuracy 0.061056 0.683410 308.000000
The first statistic that my eyes were drawn to when I saw this output was the R^2 statistic. It looks quite low and I’m not even sure why. That being said, status in the KDD Cup 2015 competition is measured in AUC, and here you can see that it is .75 on my validation data. Next, have a look at the confusion matrix. You can see in the Error column that the model did quite well predicting who would drop out (naturally, in my opinion), but did not do so well figuring out who would stay. The overall error rate on the validation data is 10%, but I’m still not so happy about the high error rate as it pertains to those who stayed in the MOOC.
So this was all well and good (and was what got me my highest score yet according to the KDD Cup leaderboard) but what if I could get better performance with fewer variables? I took a look at my variable importances and decided to see what would happen if I eliminate the variables with the lowest importance scores one by one until I reach the variable with the 16th lowest importance score. Here’s the code I used:
varimps = data.frame(h2o.varimp(train.gbm))
variable.set = list(nvars = c(), AUC = c(), min.AUC = c(), max.AUC = c())
mooc.hex = as.h2o(localH2O, mooc[,c("enrollment_id","dropped_out_factor",x.names)])
n = 1
for (i in seq(35,20)) {
auc.temp = c(NA,NA,NA)
x.names.new = setdiff(x.names, varimps$variable[i:dim(varimps)[1]])
for (j in 1:3) {
mooc.hex.split = h2o.splitFrame(mooc.hex, ratios=.8)
train.gbm.smaller = h2o.gbm(x = x.names.new, y = "dropped_out_factor", training_frame = mooc.hex.split[[1]],
validation_frame = mooc.hex.split[[2]], ntrees = 500, balance_classes = FALSE, learn_rate = .05)
auc.temp[j] = train.gbm.smaller@model$validation_metrics@metrics$AUC
}
variable.set$AUC[n] = mean(auc.temp)
variable.set$min.AUC[n] = min(auc.temp)
variable.set$max.AUC[n] = max(auc.temp)
variable.set$nvars[n] = i-1
n = n + 1
}
variable.set.df = data.frame(variable.set)
You can see that it’s a similar algorithm as what I used to do the model tuning. I moved up the variable importance list from the bottom, one variable at a time, and progressively eliminated more variables. I trained 3 models for each new number of variables, each on a random sample of the data, and averaged the AUCs from those models (totalling 48 models). See the following graph for the result:
As you can see, even though the variables I eliminated were of the lowest importance, they were still contributing something positive to the model. This goes to show how well GBM performs with variables that could be noisy.
Now let’s look at the more important variables according to H2O:
variable relative_importance scaled_importance percentage
1 num_logged_events 48481.160156 1.000000e+00 5.552562e-01
2 DAYS_problem_total_etime_unix 11651.416992 2.403288e-01 1.334440e-01
3 days.in.mooc 6495.756348 1.339852e-01 7.439610e-02
4 DAYS_access_total_etime_unix 3499.054443 7.217349e-02 4.007478e-02
5 avg_month 3019.399414 6.227985e-02 3.458127e-02
6 avg_day 1862.299316 3.841285e-02 2.132897e-02
7 Pct_sequential 1441.578247 2.973481e-02 1.651044e-02
8 DAYS_navigate_total_etime_unix 969.427734 1.999597e-02 1.110289e-02
9 num_courses 906.499451 1.869797e-02 1.038217e-02
10 Pct_problem 858.774353 1.771357e-02 9.835569e-03
11 num_students 615.350403 1.269257e-02 7.047627e-03
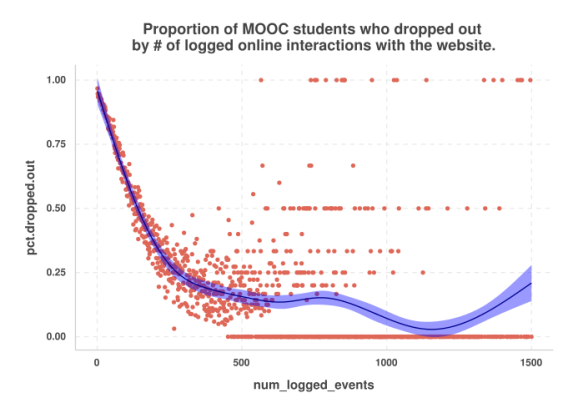
Firstly, we see that the number of logged events was the most important variable for predicting drop-out. I guess the more active they are, the less likely they are to drop out. Let’s see a graph:
Although a little bit messy because I did not bin the num_logged_events variable, we see that this is exactly the case that those students who were more active online were less likely to drop out.
Next, we see a few variables regarding the days spent doing something. They seem to follow similar patterns, so the image I’ll show you below involves the days.in.mooc variable. This is simply how many days passed from the logging of the first event to the last.
Here we see a very steady decrease in probability of dropping out where those who spent very little time from their first to their last interaction with the MOOC are the most likely to drop out, whereas those who spend more time with it are obviously less likely.
Next, let’s look at the avg_month and avg_day variables. These were calculated by taking the average timestamp of all events for each person enrolled in each course and then extracting the month and then the day from that timestamp. Essentially, when, on average, did they tend to do that course.
Interestingly, most months seem to exhibit a downward pattern, whereby if the person tended to have their interactions with the MOOC near the end of the month, then they were less likely to drop out, but if they had their interactions near the beginning they were more likely to drop out. This applied to February, May, June, November, and December. The reverse seems to be true for July and maybe October. January maybe applies to the second list.
The last two plots I’ll show you relate to num_courses and num_students, in other words, how many courses each student is taking and how many students are in each course.
The interesting result here is that it’s only those students who were super committed (taking more than 20 courses in the period captured by the data) who appeared significantly less likely to drop out than those who were taking fewer courses.
Finally, you can see that as the number of students enrolled in a course went up, the overall drop-out rate decreased. Popular courses retain students!
Conclusion
This was fun! I was amazed by how obsessed I became on account of this competition. I’m disappointed that I couldn’t think of something to bridge the 2.6% gap between me and first place, but the point of this was to practice, to learn something new, and have fun. I hope you enjoyed it too!
R-bloggers.com offers daily e-mail updates about R news and tutorials on topics such as: visualization (ggplot2, Boxplots, maps, animation), programming (RStudio, Sweave, LaTeX, SQL, Eclipse, git, hadoop, Web Scraping) statistics (regression, PCA, time series, trading) and more...